Radiology Initiatives Illustrate Uses for Open Data and Open AI research
 Andy OramFans of data in health care often speculate about what clinicians and researchers could achieve by reducing friction in data sharing. What if we had easy access to group repositories, expert annotations and labels, robust and consistent metadata, and standards without inconsistencies? Since 2017, the Radiological Society of North America (RSNA) has been displaying a model for such data sharing.
Andy OramFans of data in health care often speculate about what clinicians and researchers could achieve by reducing friction in data sharing. What if we had easy access to group repositories, expert annotations and labels, robust and consistent metadata, and standards without inconsistencies? Since 2017, the Radiological Society of North America (RSNA) has been displaying a model for such data sharing.
That year marked RSNA's first AI challenge. RSNA has worked since then to make the AI challenge an increasingly international collaboration. Organizers of each challenge curate and annotate medical imaging studies and ask the research community to come up with models to answer important questions. I talked about the RSNA challenges and broader AI data collection efforts with Matthew P. Lungren, MD, MPH, who is widely recognized for his work on AI in radiology and is co-director of the Stanford Center for Artificial Intelligence in Medicine and Imaging.
 Matthew P. Lungren, MD, MPHTwo things have to come together for effective AI nowadays: large data sets in a common format and accurate annotations or labels for the data. These ingredients require experienced clinicians and researchers who can identify clinically important imaging problems, curate large imaging datasets, and produce expert annotations for the data–all to “teach” AI algorithms to learn these human expert tasks. We'll take a close look at the data characteristics that make the RSNA AI data challenges successful.
Matthew P. Lungren, MD, MPHTwo things have to come together for effective AI nowadays: large data sets in a common format and accurate annotations or labels for the data. These ingredients require experienced clinicians and researchers who can identify clinically important imaging problems, curate large imaging datasets, and produce expert annotations for the data–all to “teach” AI algorithms to learn these human expert tasks. We'll take a close look at the data characteristics that make the RSNA AI data challenges successful.
Start with a standard—not the porous, fuzzy standards so prevalent in health care, but the DICOM standard released in 1992. DICOM is truly interoperable; images produced by different machines from different manufacturers can be read and compared smoothly. DICOM also stores important non-imaging metadata about the patient and the examination in HIPAA-compliant fashion. RSNA taps this information when curating multi-institutional datasets. Common DICOM deidentification techniques are important to protect patients. Institutions donating the images can use a DICOM Anonymizer provided by RSNA, or their own anonymizing techniques.
Reliable diagnostic information is also crucial. Most AI models are trained on data curated and labeled to address a specific question or diagnosis. That is, you start with a question such as "What does a brain hemorrhage look like?" You then take a large set of images (hundreds, and hopefully even thousands), some of which show brain hemorrhages and others that don't.
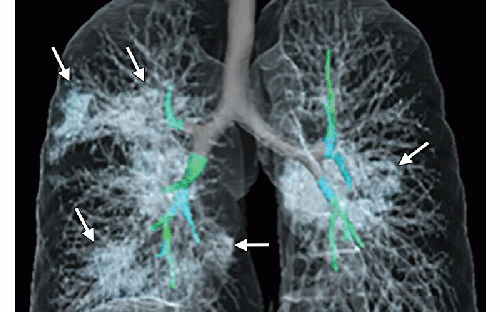 Images in a 41-year-old woman who presented with fever and positive polymerase chain reaction assay for the 2019 novel coronavirus (2019-nCoV). Three representative axial thin-section chest CT images show multifocal ground glass opacities without consolidation. Three-dimensional volume-rendered reconstruction shows the distribution of the ground-glass opacities (arrows). Image courtesy of the Radiological Society of North America (RSNA)
Images in a 41-year-old woman who presented with fever and positive polymerase chain reaction assay for the 2019 novel coronavirus (2019-nCoV). Three representative axial thin-section chest CT images show multifocal ground glass opacities without consolidation. Three-dimensional volume-rendered reconstruction shows the distribution of the ground-glass opacities (arrows). Image courtesy of the Radiological Society of North America (RSNA)
The labels are typically provided by expert radiologists. Researchers train a model to recognize hemorrhages in brain scans, then test the model on a smaller set of images that were not used to train the model. This training procedure can tolerate a certain amount of noise, including incorrectly diagnosed images, but it depends on most of them being accurately identified.
So the RSNA sends every image in the challenge to one or more radiologists for analysis and annotation. A diverse dataset consisting of imaging studies from many hospitals and patient populations are important so that the AI model does not “learn” to recognize the abnormality on only one type of scanner or patient population, but is instead able to recognize the abnormality in many possible clinical settings.
Images currently come from four countries, and the RSNA will add more in upcoming challenges. Completed challenges include:
- Identifying the age of children's bones (2017, 37 participating teams)
- Detecting pneumonia from chest X-rays (2018, 1,400 participating teams)
- Detecting intracranial hemorrhages from cranial CT exams (2019, 1,345 participating teams)
For 2020, the RSNA used its extensive network and its experience at building large annotated datasets from earlier challenges to create a COVID-19 data collection forAI researchers. This effort, led by RSNA in combination with The Cancer Imaging Archive, is creating a data set with chest CT exams and X-rays, called the RSNA International COVID-19 Open Radiology Database (RICORD). The research objectives are broader than the challenges and will make the annotated COVID-19 images data available to all researchers to answer questions about COVID-19. More than 200 institutions around the world have expressed their interest in participating.
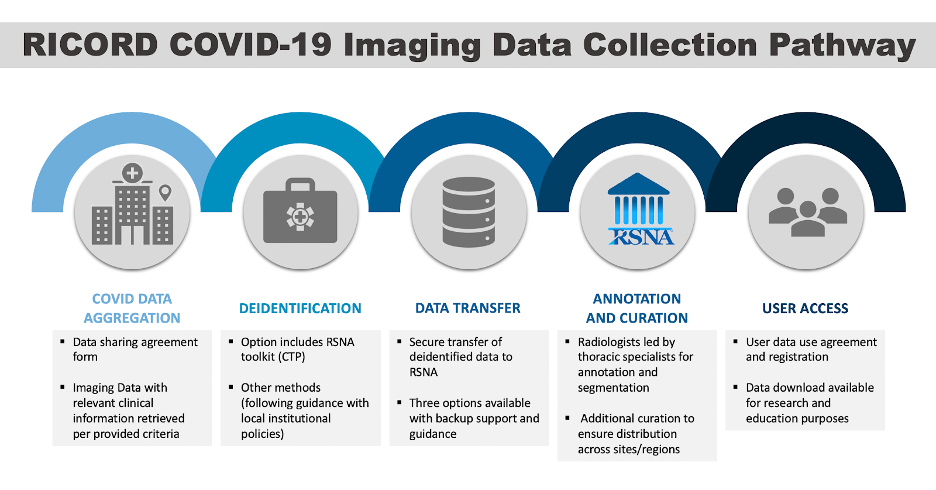 Lungren says that at the end of each challenge, many participants release their models as open source. This means access to the results of their research by all clinical and research facilities in the world. The RICORD dataset effort will also ideally lead to more open source AI models as well. The experience with international medical AI data challenges has laid a foundation for the application of shared data to health care.
Lungren says that at the end of each challenge, many participants release their models as open source. This means access to the results of their research by all clinical and research facilities in the world. The RICORD dataset effort will also ideally lead to more open source AI models as well. The experience with international medical AI data challenges has laid a foundation for the application of shared data to health care.
"More than ever, this pandemic is showing us that we can rally together toward a common purpose," Dr. Lungren said. "Rather than siloing data and pursuing fractured efforts, we can instead choose to collaborate through efforts like RICORD to accelerate an end to this pandemic as a united global imaging community."
In the announcement of the launching of RICORD, RSNA acknowledged the support of the following societies in helping to bring global imaging data to the repository: Asian Oceanian Society of Radiology, Japan Radiological Society, Federación Argentina de Asociaciones de Radiología, Diagnóstico por Imágenes y Terapia Radiante, Sociedad Argentina de Radiología, Sociedad Chilena de Radiología, Sociedade Paulista de Radiologia e Diagnóstico por Imagem, European Society of Medical Imaging Informatics, and Netherlands Cancer Institute.
Sites interested in learning more or contributing data should visit the RICORD resources page. Additional RSNA COVID-19 resources may be found at RSNA.org/COVID-19.
- Tags:
- 2019 novel coronavirus (2019-nCoV)
- accurate annotations for the data
- accurate labels for the data
- AI algorithms
- AI data collection efforts
- Andy Oram
- annotate medical imaging studies
- annotated COVID-19 images data
- application of shared data to health care
- Asian Oceanian Society of Radiology
- clinically important imaging problems
- consistent metadata
- consistent standards
- COVID-19
- COVID-19 data collection for AI researchers
- curate medical imaging studies
- curating large imaging datasets
- curating multi-institutional datasets
- data sharing
- DICOM Anonymizer
- DICOM deidentification techniques
- DICOM interoperability
- DICOM standard
- DICOM storage of non-imaging metadata
- European Society of Medical Imaging Informatics
- expert annotations
- expert labels
- Federación Argentina de Asociaciones de Radiología
- global imaging data repository
- group repositories
- health care
- HIPAA compliance
- imaging studies
- international collaboration
- international medical AI data challenges
- Japan Radiological Society
- large annotated datasets
- large data sets in a common
- Matthew P. Lungren
- multifocal ground glass opacities
- Netherlands Cancer Institute
- Open AI
- Open Data
- open health
- open source
- open source AI models
- Polymerase chain reaction (PCR)
- Radiological Society of North America (RSNA)
- radiologists
- reducing friction in data sharing
- RSNA AI challenge
- RSNA International COVID-19 Open Radiology Database (RICORD)
- Sociedad Argentina de Radiología
- Sociedad Chilena de Radiología
- Sociedade Paulista de Radiologia
- Stanford Center for Artificial Intelligence in Medicine and Imaging
- The Cancer Imaging Archive
- Login to post comments